
Education
- University of Kansas, BS Biology, 1997
- University of Texas at Austin, Ph.D Molecular Biology, 2005
- University of Texas at Austin, Research Associate, 2005-2009
Professional Experience
- Associate Professor, 2017-present, Department of Chemistry and Biochemistry, University of Maryland, College Park
- Assistant Professor, 2010-2017, Department of Chemistry and Biochemistry, University of Maryland, College Park
Research Interests
Nucleic acid structure, function, and catalysis. Structural DNA nanotechnology.
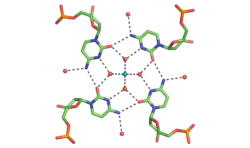 Determining the double helical structure of DNA was one of the most important scientific achievements of the 20th century. The predictability of G-C and A-T base pairs that are necessary for its biological functions have also made DNA one of the most widely used molecules for the construction of nanoscale objects. Despite this, there are limitations to the Watson-Crick duplex. My laboratory is pushing the beyond the traditional Watson and Crick boundaries of DNA self-assembly. Using x-ray crystallography and other biophysical techniques we are focused on finding new DNA motifs, understanding their characteristics, and using them for construction of DNA assemblies.
Determining the double helical structure of DNA was one of the most important scientific achievements of the 20th century. The predictability of G-C and A-T base pairs that are necessary for its biological functions have also made DNA one of the most widely used molecules for the construction of nanoscale objects. Despite this, there are limitations to the Watson-Crick duplex. My laboratory is pushing the beyond the traditional Watson and Crick boundaries of DNA self-assembly. Using x-ray crystallography and other biophysical techniques we are focused on finding new DNA motifs, understanding their characteristics, and using them for construction of DNA assemblies.
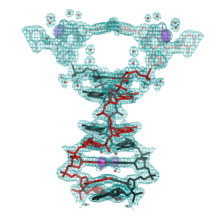 We have described a number of previously unobserved DNA motifs. We described the first example of a non-canonical DNA structural “switch” that is capable of responding to changes in pH and results in concerted movements within single crystals (Muser & Paukstelis, 2012). We described the first crystal structure a DNA G-quadruplex structure that also contains C-tetrads and barium cations (Zhang et al., 2014). We have also found an entirely new class of DNA tetraplex motifs formed by parallel-stranded DNA duplexes and nucleobase intercalation (Tripathi et al., 2015). We have also undertaken large scale crystallization efforts to understand how sequence influences DNA self-assembly into crystals (Saoji et al., 2015).
We have described a number of previously unobserved DNA motifs. We described the first example of a non-canonical DNA structural “switch” that is capable of responding to changes in pH and results in concerted movements within single crystals (Muser & Paukstelis, 2012). We described the first crystal structure a DNA G-quadruplex structure that also contains C-tetrads and barium cations (Zhang et al., 2014). We have also found an entirely new class of DNA tetraplex motifs formed by parallel-stranded DNA duplexes and nucleobase intercalation (Tripathi et al., 2015). We have also undertaken large scale crystallization efforts to understand how sequence influences DNA self-assembly into crystals (Saoji et al., 2015).
 Much of our work is geared toward being able to rationally design DNA crystals for nanotechnology applications. We described the first rationally designed 3-D DNA crystals, based on a predictable non-canonical base pairing motif. Subsequently, we have demonstrated that these crystals contain a network of solvent channels into which macromolecules can be absorbed or excluded based on size. Building on this work, we have demonstrated that protein enzymes encapsulated in the DNA crystals are active (Geng & Paukstelis, 2014), making these the first example of DNA-based solid-state biocatalysts. My laboratory is continuing to develop DNA-based technologies at the emerging interface between biology, materials science, and engineering.
Much of our work is geared toward being able to rationally design DNA crystals for nanotechnology applications. We described the first rationally designed 3-D DNA crystals, based on a predictable non-canonical base pairing motif. Subsequently, we have demonstrated that these crystals contain a network of solvent channels into which macromolecules can be absorbed or excluded based on size. Building on this work, we have demonstrated that protein enzymes encapsulated in the DNA crystals are active (Geng & Paukstelis, 2014), making these the first example of DNA-based solid-state biocatalysts. My laboratory is continuing to develop DNA-based technologies at the emerging interface between biology, materials science, and engineering.


